Planet Python
Last update: March 10, 2026 04:44 PM UTC
March 10, 2026
Real Python
Working With APIs in Python: Reading Public Data
Python is an excellent choice for working with Application Programming Interfaces (APIs), allowing you to efficiently consume and interact with them. By using the Requests library, you can easily fetch data from APIs that communicate using HTTP, such as REST, SOAP, or GraphQL APIs. This video course covers the essentials of consuming REST APIs with Python, including authentication and handling responses.
By the end of this video course, you’ll understand that:
- An API is an interface that allows different systems to communicate, typically through requests and responses.
- Python is a versatile language for consuming APIs, offering libraries like Requests to simplify the process.
- REST and GraphQL are two common types of APIs, with REST being more widely used for public APIs.
- To handle API authentication in Python, you can use API keys or more complex methods like OAuth to access protected resources.
Knowing how to consume an API is one of those magical skills that, once mastered, will crack open a whole new world of possibilities, and consuming APIs using Python is a great way to learn such a skill.
By the end of this video course, you’ll be able to use Python to consume most of the APIs that you come across. If you’re a developer, then knowing how to consume APIs with Python will empower you to integrate data from various online sources into your applications.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Quiz: Python Descriptors: An Introduction
In this quiz, you’ll test your understanding of Python Descriptors.
By working through this quiz, you’ll revisit the descriptor protocol, how .__get__() and .__set__() control attribute access, and how to implement read only descriptors. You’ll also explore data vs. non-data descriptors, attribute lookup order, and the .__set_name__() method.
These exercises help you reason about real descriptor behavior and see when and why to use them in your code.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Wingware
Wing Python IDE Version 11.1 - March 10, 2026
Wing Python IDE version 11.1 has been released with improved pseudo TTY emulation in OS Commands, Start Terminal on Windows, syntax highlighting for Rust and TOML, support for newer OpenAI models in AI Chat, new Stop on SystemExit debugger preference, and other minor features and bug fixes.
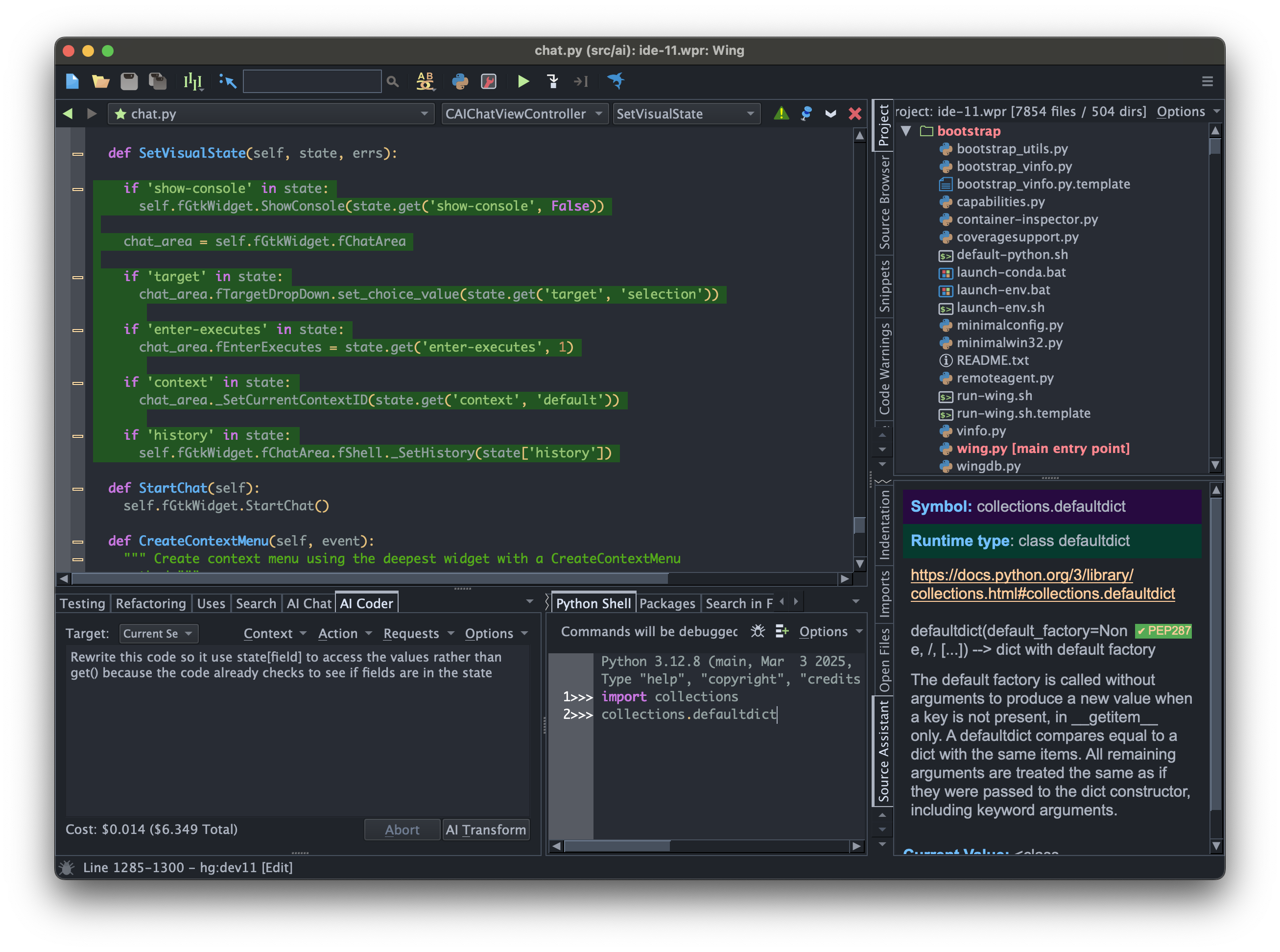
Downloads
Wing 10 and earlier versions are not affected by installation of Wing 11 and may be installed and used independently. However, project files for Wing 10 and earlier are converted when opened by Wing 11 and should be saved under a new name, since Wing 11 projects cannot be opened by older versions of Wing.
 New in Wing 11
New in Wing 11
Improved AI Assisted Development
Wing 11 improves the user interface for AI assisted development by introducing two separate tools AI Coder and AI Chat. AI Coder can be used to write, redesign, or extend code in the current editor. AI Chat can be used to ask about code or iterate in creating a design or new code without directly modifying the code in an editor.
Wing 11's AI assisted development features now support not just OpenAI but also Claude, Grok, Gemini, Perplexity, Mistral, Deepseek, and any other OpenAI completions API compatible AI provider.
This release also improves setting up AI request context, so that both automatically and manually selected and described context items may be paired with an AI request. AI request contexts can now be stored, optionally so they are shared by all projects, and may be used independently with different AI features.
AI requests can now also be stored in the current project or shared with all projects, and Wing comes preconfigured with a set of commonly used requests. In addition to changing code in the current editor, stored requests may create a new untitled file or run instead in AI Chat. Wing 11 also introduces options for changing code within an editor, including replacing code, commenting out code, or starting a diff/merge session to either accept or reject changes.
Wing 11 also supports using AI to generate commit messages based on the changes being committed to a revision control system.
You can now also configure multiple AI providers for easier access to different models.
For details see AI Assisted Development under Wing Manual in Wing 11's Help menu.
Package Management with uv
Wing Pro 11 adds support for the uv package manager in the New Project dialog and the Packages tool.
For details see Project Manager > Creating Projects > Creating Python Environments and Package Manager > Package Management with uv under Wing Manual in Wing 11's Help menu.
Improved Python Code Analysis
Wing 11 makes substantial improvements to Python code analysis, with better support for literals such as dicts and sets, parametrized type aliases, typing.Self, type of variables on the def or class line that declares them, generic classes with [...], __all__ in *.pyi files, subscripts in typing.Type and similar, type aliases, type hints in strings, type[...] and tuple[...], @functools.cached_property, base classes found also in .pyi files, and typing.Literal[...].
Updated Localizations
Wing 11 updates the German, French, and Russian localizations, and introduces a new experimental AI-generated Spanish localization. The Spanish localization and the new AI-generated strings in the French and Russian localizations may be accessed with the new User Interface > Include AI Translated Strings preference.
Improved diff/merge
Wing Pro 11 adds floating buttons directly between the editors to make navigating differences and merging easier, allows undoing previously merged changes, and does a better job managing scratch buffers, scroll locking, and sizing of merged ranges.
For details see Difference and Merge under Wing Manual in Wing 11's Help menu.
Other Minor Features and Improvements
Wing 11 also adds support for Python 3.14, improves the custom key binding assignment user interface, adds a Files > Auto-Save Files When Wing Loses Focus preference, warns immediately when opening a project with an invalid Python Executable configuration, allows clearing recent menus, expands the set of available special environment variables for project configuration, and makes a number of other bug fixes and usability improvements.
Changes and Incompatibilities
Since Wing 11 replaced the AI tool with AI Coder and AI Chat, and AI configuration is completely different than in Wing 10, you will need to reconfigure your AI integration manually in Wing 11. This is done with Manage AI Providers in the AI menu. After adding the first provider configuration, Wing will set that provider as the default. You can switch between providers with Switch to Provider in the AI menu.
If you have questions, please don't hesitate to contact us at support@wingware.com.
March 09, 2026
The Python Coding Stack
Field Notes: First, Second, and Five Hundred and Twenty-Third • [Club]
Most of my posts on The Python Coding Stack, whether in the main publication or here in The Club, typically focus on some aspect of core Python and explore it through a step-by-step approach, a mini-project, or sometimes through an essay-type article.
But today, I’ll write a short post about some tools I came across that may be interesting. I’ll keep the post short since, if you’re interested, you can easily explore the packages independently – you won’t need my explanatory efforts.
Remember some of the earliest code you ever wrote? It may have looked like this:
first = input(”Enter the first number: “)
second = input(”Enter the second number: “)
print(
f”The sum of the two numbers is {float(first) + float(second)}”
)Those days are long gone. But what if you wanted this code to work for any number of inputs, not just two?
I’m not talking about the code to work out the sum itself. You’d probably use a list to collect all the numbers and the sum() built-in function, or just use a running total. Whatever.
The annoying part is making user-friendly strings when asking for the input – it’s fine to write "first" and "second" when there are only two prompts – and then when showing the result, which currently says "two numbers". But what if you have more?
I came across this need several times, but my solution has always been to change the string so that it’s general enough to work in all cases. I’m lazy, I know.
In any case, recently I decided to look for solutions. And of course, they exist! Everything seems to exist in the Python ecosystem. Several solutions…
num2words
The first solution is probably the simplest: the num2words package doesn’t do much beyond converting numbers to words, as its name suggests. You’ll need to install num2words using pip, uv, or whatever you use to install packages.
>>> import num2words
>>> num2words.num2words(34)
‘thirty-four’The main function in the num2words module is also called num2words(), as often happens with such modules!
How far can it go?
>>> num2words.num2words(5468.23)
‘five thousand, four hundred and sixty-eight point two three’Working in Spanish?
>>> num2words.num2words(5468.23, lang=”es”)
‘cinco mil cuatrocientos sesenta y ocho punto dos tres’Or Japanese?
>>> num2words.num2words(5468.23, lang=”ja”)
‘五千四百六十八点二三’Real Python
Python Gains frozendict and Other Python News for March 2026
After years of community requests, Python is finally getting frozendict. The Steering Council accepted PEP 814 in February, bringing an immutable, hashable dictionary as a built-in type in Python 3.15. It’s one of those additions that feels overdue, and the frozenset-to-set analogy makes it immediately intuitive. This is just one piece of a busy month of Python news.
Beyond that, February brought security patches, AI SDK updates, and some satisfying infrastructure improvements under Python’s hood. Time to dive into the biggest Python news from the past month!
Join Now: Click here to join the Real Python Newsletter and you’ll never miss another Python tutorial, course, or news update.
Python Releases and PEP Highlights
The Steering Council was active in February, with several PEP decisions landing. On the release side, both the 3.14 and 3.13 branches got maintenance updates.
Python 3.15.0 Alpha 6: Comprehension Unpacking and More
Python 3.15.0a6 shipped on February 11, continuing the alpha series toward the May 5 beta freeze. This release includes several accepted PEPs that are now testable:
- PEP 798: Unpacking in comprehensions using
*and** - PEP 799: A high-frequency, low-overhead statistical sampling profiler
- PEP 686: UTF-8 as the default text encoding
- PEP 728:
TypedDictwith typed extra items
PEP 798 is the kind of quality-of-life improvement that makes you smile. It lets you flatten or merge collections directly in a comprehension:
>>> lists = [[1, 2], [3, 4], [5]]
>>> [*it for it in lists]
[1, 2, 3, 4, 5]
>>> dicts = [{"a": 1}, {"b": 2}]
>>> {**d for d in dicts}
{'a': 1, 'b': 2}
No more writing explicit loops just to concatenate a list of lists.
The JIT compiler continues to show gains: 3-4% on x86-64 Linux over the standard interpreter and 7-8% on AArch64 macOS over the tail-calling interpreter, matching the numbers from alpha 5.
Note: Alpha 7 is scheduled for March 10, 2026, with the beta phase starting May 5. If you maintain packages, now is a great time to test against early builds.
Python 3.14.3 and 3.13.12: Maintenance Releases
On February 3, the team shipped Python 3.14.3 with around 300 bug fixes and Python 3.13.12 with about 240 fixes. No new features here, but if you’re running either version in production, it’s worth grabbing these patches to stay current.
PEP 814 Accepted: frozendict Joins the Built-Ins
This one has been on many Python developers’ wishlists for over a decade. PEP 814, authored by Victor Stinner and Donghee Na, adds frozendict as a built-in immutable dictionary type in Python 3.15.
The concept is straightforward. Just as frozenset gives you an immutable version of set, frozendict gives you an immutable version of dict:
>>> config = frozendict(host="localhost", port=8080)
>>> config["host"]
'localhost'
>>> config["host"] = "0.0.0.0"
Traceback (most recent call last):
...
TypeError: 'frozendict' object does not support item assignment
Read the full article at https://realpython.com/python-news-march-2026/ »
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Quiz: Introduction to Python SQL Libraries
In this quiz, you’ll test your understanding of Python SQL Libraries.
Work through this quiz to connect to databases with Python, interact with SQLite, MySQL, and PostgreSQL, run SQL queries, and write scripts that work across databases.
Reinforce your practical skills for building Python apps backed by databases.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Quiz: Pydantic AI: Build Type-Safe LLM Agents in Python
In this quiz, you’ll test your understanding of Pydantic AI: Build Type-Safe LLM Agents in Python.
You’ll revisit what Pydantic AI is, how to specify a model provider, how to return structured outputs using Pydantic models, how to register and select tools, how dependency injection works, and what trade-offs to expect in production.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Python Bytes
#472 Monorepos
<strong>Topics covered in this episode:</strong><br> <ul> <li><strong><a href="https://dev.to/aws/3-things-i-wish-i-knew-before-setting-up-a-uv-workspace-30j6?featured_on=pythonbytes">Setting up a Python monorepo with uv workspaces</a></strong></li> <li><strong><a href="https://catt.rs/en/stable/?featured_on=pythonbytes">cattrs</a>: Flexible Object Serialization and Validation</strong></li> <li><strong><a href="https://jblanca.net/edu/learning_programming_in_ai_age/?featured_on=pythonbytes">Learning to program in the AI age</a></strong></li> <li><strong><a href="https://www.linkedin.com/posts/savannahostrowski_hey-you-are-you-using-fastapi-well-activity-7432877782914977793-2ayP/?utm_medium=ios_app&rcm=ACoAAABOjqABPkOWTTbZXV9tmnQohvpkplQOibU&utm_source=social_share_send&utm_campaign=share_via&featured_on=pythonbytes">VS Code extension</a> for FastAPI and friends</strong></li> <li><strong>Extras</strong></li> <li><strong>Joke</strong></li> </ul><a href='https://www.youtube.com/watch?v=eeT0gRrQVmU' style='font-weight: bold;'data-umami-event="Livestream-Past" data-umami-event-episode="472">Watch on YouTube</a><br> <p><strong>About the show</strong></p> <p>Sponsored by us! Support our work through:</p> <ul> <li>Our <a href="https://training.talkpython.fm/?featured_on=pythonbytes"><strong>courses at Talk Python Training</strong></a></li> <li><a href="https://courses.pythontest.com/p/the-complete-pytest-course?featured_on=pythonbytes"><strong>The Complete pytest Course</strong></a></li> <li><a href="https://www.patreon.com/pythonbytes"><strong>Patreon Supporters</strong></a> <strong>Connect with the hosts</strong></li> <li>Michael: <a href="https://fosstodon.org/@mkennedy">@mkennedy@fosstodon.org</a> / <a href="https://bsky.app/profile/mkennedy.codes?featured_on=pythonbytes">@mkennedy.codes</a> (bsky)</li> <li>Brian: <a href="https://fosstodon.org/@brianokken">@brianokken@fosstodon.org</a> / <a href="https://bsky.app/profile/brianokken.bsky.social?featured_on=pythonbytes">@brianokken.bsky.social</a></li> <li>Show: <a href="https://fosstodon.org/@pythonbytes">@pythonbytes@fosstodon.org</a> / <a href="https://bsky.app/profile/pythonbytes.fm">@pythonbytes.fm</a> (bsky) Join us on YouTube at <a href="https://pythonbytes.fm/stream/live"><strong>pythonbytes.fm/live</strong></a> to be part of the audience. Usually <strong>Monday</strong> at 11am PT. Older video versions available there too. Finally, if you want an artisanal, hand-crafted digest of every week of the show notes in email form? Add your name and email to <a href="https://pythonbytes.fm/friends-of-the-show">our friends of the show list</a>, we'll never share it.</li> </ul> <p><strong>Brian #1: <a href="https://dev.to/aws/3-things-i-wish-i-knew-before-setting-up-a-uv-workspace-30j6?featured_on=pythonbytes">Setting up a Python monorepo with uv workspaces</a></strong></p> <ul> <li>Dennis Traub</li> <li>The 3 things <ul> <li>Give the Root a Distinct Name</li> <li>Use workspace = true for Inter-Package Deps</li> <li>Use importlib Mode for pytest</li> </ul></li> </ul> <p><strong>Michael #2: <a href="https://catt.rs/en/stable/?featured_on=pythonbytes">cattrs</a>: Flexible Object Serialization and Validation</strong></p> <ul> <li><strong>cattrs</strong> is a Swiss Army knife for (un)structuring and validating data in Python.</li> <li>A natural alternative/follow on from <a href="https://github.com/rnag/dataclass-wizard?featured_on=pythonbytes">DataClass Wizard</a></li> <li>Converts to ←→ from dictionaries</li> <li><em>cattrs</em> also focuses on <strong>functional composition</strong> and <strong>not coupling</strong> your data model to its serialization and validation rules.</li> <li>When you’re handed unstructured data (by your network, file system, database, …), <em>cattrs</em> helps to convert this data into trustworthy structured data.</li> <li>Batteries Included: cattrs comes with pre-configured converters for a number of serialization libraries, including JSON (standard library, orjson, UltraJSON), msgpack, cbor2, bson, PyYAML, tomlkit and msgspec (supports only JSON at this time).</li> </ul> <p><strong>Brian #3: <a href="https://jblanca.net/edu/learning_programming_in_ai_age/?featured_on=pythonbytes">Learning to program in the AI age</a></strong></p> <ul> <li>Jose Blanca</li> <li>“I teach a couple of introductory Python courses and I've been thinking about which advice to give to my students, that are studying how to program for the first time. I have collected my ideas in these blog posts” <ul> <li><a href="https://jblanca.net/blog/2026/03/06/learning-to-code-in-the-ai-age/?featured_on=pythonbytes">Why</a> learning to program is as useful as ever, even with powerful AI tools available.</li> <li><a href="https://jblanca.net/blog/2026/03/07/the-art-of-learning-in-the-ai-age/?featured_on=pythonbytes">How</a> to use AI as a tutor rather than a shortcut, and why practice remains the key to real understanding.</li> <li><a href="https://jblanca.net/blog/2026/03/08/programming-what-to-learn/?featured_on=pythonbytes">What</a> the real learning objectives are: mental models, managing complexity, and thinking like a software developer.</li> </ul></li> </ul> <p><strong>Michael #4: <a href="https://www.linkedin.com/posts/savannahostrowski_hey-you-are-you-using-fastapi-well-activity-7432877782914977793-2ayP/?utm_medium=ios_app&rcm=ACoAAABOjqABPkOWTTbZXV9tmnQohvpkplQOibU&utm_source=social_share_send&utm_campaign=share_via&featured_on=pythonbytes">VS Code extension</a> for FastAPI and friends</strong></p> <ul> <li>Enhances the FastAPI development experience in Visual Studio Code</li> <li><strong>Path Operation Explorer</strong>: Provides a hierarchical tree view of all FastAPI routes in your application.</li> <li><strong>Search for routes</strong>: Use the Command Palette and quickly search for routes by path, method, or name.</li> <li><strong>CodeLens links appear above HTTP client calls</strong> like client.get('/items'), letting you jump directly to the matching route definition.</li> <li><strong>Deploy your application</strong> directly to <a href="https://fastapicloud.com/?featured_on=pythonbytes">FastAPI Cloud</a> from the status bar with zero config.</li> <li>View <strong>real-time logs from your FastAPI Cloud</strong> deployed applications directly within VS Code.</li> <li><a href="https://marketplace.visualstudio.com/items?itemName=FastAPILabs.fastapi-vscode&featured_on=pythonbytes">Install from Marketplace</a>.</li> </ul> <p><strong>Extras</strong></p> <p>Brian:</p> <ul> <li><a href="https://gvanrossum.github.io/interviews/index.html?featured_on=pythonbytes">Guido van Rossum interviews key Python developers from the first 25 years</a> <ul> <li><a href="https://gvanrossum.github.io/interviews/BrettCannon.html?featured_on=pythonbytes"><strong>Interview with Brett Cannon</strong></a></li> <li><a href="https://gvanrossum.github.io/interviews/ThomasWouters.html?featured_on=pythonbytes"><strong>Interview with Thomas Wouters</strong></a> Michael:</li> </ul></li> <li><a href="https://www.youtube.com/watch?v=Kourq_Lz03U">IntelliJ IDEA: The Documentary | An origin story</a> video</li> <li>Cursor Joined <a href="https://www.jetbrains.com/acp/?featured_on=pythonbytes">the ACP Registry</a> and Is <a href="https://blog.jetbrains.com/ai/2026/03/cursor-joined-the-acp-registry-and-is-now-live-in-your-jetbrains-ide/?utm_source=marketo&utm_medium=email&utm_content=blog&utm_campaign=ai&lidx=0&wpid=685326&mkt_tok=NDI2LVFWRC0xMTQAAAGgXJQdteprtfw26Lw-urwLQEjJcm8qYIOseeX-9jODplxTohhXyxl11ZbYr2ehMl__lEyRq0YT8B-mqpzj-fMmOeHXBN0Lz1o9_QOCmx7qSij-fDKY&featured_on=pythonbytes">Now Live in Your JetBrains IDE</a></li> <li><a href="https://mkennedy.codes/posts/what-hyper-personal-software-looks-like/?featured_on=pythonbytes">What hyper-personal software looks like</a></li> <li>I’m doing in-person training again (limited scope): <ul> <li><a href="https://mkennedy.codes/agentic-ai-enablement/?featured_on=pythonbytes">On-site, hands-on AI engineering enablement for software teams with Michael</a></li> </ul></li> </ul> <p>Joke: <a href="https://www.reddit.com/r/SaasDevelopers/comments/1rcdzrk/saas_is_dead/?share_id=Zi4lrnVSFkytCUnpfYIMo&utm_content=1&utm_medium=ios_app&utm_name=ioscss&utm_source=share&utm_term=1&featured_on=pythonbytes">Saas is dead</a></p>
Mariatta
Year of the Snake Recap
Looking Back at the Year of the Snake
It’s already March 2026, you might think it’s too late for anybody to share a recap of the year 2025. Well, the good news is that we are still in the beginnings of Lunar New Year. So instead of recap of 2025, let’s review my Year of the Snake instead.
Last year was fun. I felt I was super busy with all the things, talks, conferences, but overall I felt good about it all. Even though I spent a lot of time with the Python community, I also made sure to spend quality time with my family and friends offline.
Python Anywhere
How PythonAnywhere Became a Publish Target for BeeWare Apps
tl;dr
You can now deploy BeeWare apps as web apps on
PythonAnywhere with a single command. Install the
pythonanywhere-briefcase-plugin,
run briefcase publish web static, done. There’s a
step-by-step tutorial if you want to
try it right now.
March 08, 2026
Django Weblog
DSF member of the month - Theresa Seyram Agbenyegah
For March 2026, we welcome Theresa Seyram Agbenyegah as our DSF member of the month! ⭐
Theresa is a passionate community builder serving in the DSF Events Support Working Group. She has demonstrated strong leadership by taking on roles such as LOC Programmes Lead at PyCon Africa 2024 and Programs Chair for PyCon Ghana 2025. She also organized DjangoGirls events across multiple PyCons, including PyCon Ghana 2022 and PyCon Africa 2024.
You can learn more about Theresa by visiting Theresa's LinkedIn profile and her GitHub Profile.
Let’s spend some time getting to know Theresa better!
Can you tell us a little about yourself (hobbies, education, etc)?
I’m Theresa Seyram Agbenyegah, mostly referred to in the community as Stancy; a backend engineer, social entrepreneur, and an open source advocate/contributor passionate about using technology for impact. My background is in technology, community management, and systems design. Over the years, I have grown into roles that combine engineering, leadership, and ecosystem building.
I know many folks call you Stancy, me included, why specifically this name?
So “Stancy” is my initials 😁, People think it is my nickname.
How did you start using Django?
I was introduced to Django through a Django Girls workshop, and oh i’m a Django girl. I loved how opinionated yet flexible it was. The “batteries-included” philosophy made backend architecture feel structured without being restrictive.
The admin interface especially blew my mind early on; being able to scaffold powerful internal tools so quickly felt magical.
What other frameworks do you know, and if you had magical powers, what would you add to Django?
I have worked with Flask, FastAPI, and explored the Dart framework. Each has strengths, especially FastAPI in performance and modern async patterns.
If I had magical powers, I would:
- Make async patterns even more seamless across the ecosystem
- Improve first class support for large scale distributed system
- Provide even more built-in tooling for observability and performance profiling
But overall, Django’s maturity and ecosystem are hard to beat.
What projects are you working on now?
I’m not working on any big projects at the moment, I'm mostly working on client projects at work.
Which Django libraries are your favorite (core or 3rd party)?
Some of my favorites:
- Django Rest Framework (it’s practically essential for modern APIs)
- django-filter
- django-allauth
- Celery (for async task processing)
- Django Debug Toolbar (for development clarity)
The ecosystem really makes Django powerful.
What are the top three things in Django that you like?
- The admin interface
- The ORM
- The strong community and documentation (FYI: it gives me a sense of belonging)
Django feels stable, mature, and production-ready which builds developer confidence.
You have been in the organization of PyCon Africa and DjangoGirls that happen during this conference in 2024. That's great, do you have any advice for people who would like to join or create their own DjangoGirls event in their city?
Start small and start with intention.
You don’t need a massive budget. What you need is:
- A committed small team
- Clear structure
- Support from the global DjangoGirls organization, Django Software Foundation, and other communities.
- A safe, welcoming environment
Most importantly, center the participants. The goal isn’t just teaching Django, it’s building confidence and introducing them to the Tech industry.
How did you become a leader of the PyLadies Ghana chapter?
My Leadership journey in the PyLadies Ghana community began with a simple step: attending a Django Girls workshop at Ho while I was in school. At the time, I was just curious and eager to learn more about programming. After the workshop, I was introduced to the PyLadies Ghana community and added to the group. That was my first real connection to a tech community.
I started by simply showing up, participating in conversations, attending events, and learning from others in the community. Over time, I became more involved. I joined the PyLadies Ghana Tema Chapter, where I supported the community lead with organizing activities that are bootcamps, meetups,etc. Through that experience, I had the opportunity to contribute more actively.
Because of my commitment and willingness to help, I was later asked to volunteer as a co-lead of PyLadies Ghana Tema Chapter. I accepted the opportunity and began working more closely with the Lead to organize events, support members, and grow the community. It was a period of learning, collaboration, and service.
As I continued contributing and volunteering, more opportunities opened up. When there was a chance to volunteer with PyLadies Ghana programs and events, I stepped forward again and volunteered as PyLadies Ghana Programs and Events Lead. That experience eventually led to me becoming a lead.
Looking back, my journey with PyLadies Ghana has been shaped by community, consistency, and volunteering. What started as attending a workshop grew into leadership and the chance to help create opportunities for others. It reminds me that sometimes all it takes is showing up, contributing where you can, and being willing to grow with the community.
You have been organizing a lot of events in Africa, especially in Ghana. How do you envision organizing an event? Would you like additional support?
For me, events are ecosystems, not just gatherings.
Focus on:
- Clear goals and impact
- Accessibility
- Diversity of voices
- Strong logistics planning
- Follow-up community building
Yes, more funding support, institutional partnerships for internships, and long-term sponsorship pipelines would significantly help African tech communities scale sustainably.
This is the international women's day today, I'm glad to have you featured on this special day. Do you have any word to mention in relation to this?
International Women’s Day is a reminder that representation is not a trend, it's a necessity.
We need more women building systems, shaping infrastructure, leading conversations, and owning technical spaces.
And to every woman in tech: your presence is powerful. Keep building. Keep speaking. Keep leading. Keep mentoring and raising the next tech women.
What are your hobbies or what do you do when you’re not working?
When I’m not working, I’m usually reading books/articles, mentoring, watching movies or documentaries, cooking, reflecting, or exploring new ideas around technology and social impact. I also enjoy quiet strategy sessions with myself, thinking about how to build things that outlive me.
Is there anything else you’d like to say?
Technology is more than code, it's access, power, and possibility.
I hope more people see themselves not just as users of technology, but as architects of it.
Thank you for doing the interview, Stancy !
March 07, 2026
EuroPython
Humans of EuroPython: Cristián Maureira-Fredes
Ever wonder what powers EuroPython? 🐍 No it’s not coffee—It&aposs volunteers! From stage MCs to sponsor ambassadors, Wi-Fi wizards to vibe guardians, we’re the invisible threads weaving community magic. No title, no capes—just passion.
Join us in celebrating one of the humans behind the keyboard. Read our latest interview with Cristián Maureira-Fredes, co-lead of the Programme Team at EuroPython 2025.
Words are not enough to thank you, Cristián!
 Cristián Maureira-Fredes, co-lead of the Programme Team at EuroPython 2025
Cristián Maureira-Fredes, co-lead of the Programme Team at EuroPython 2025EP: What&aposs one task you handled that attendees might not realize happens behind the scenes at EuroPython?
I believe many attendees from EuroPython and other conferences think that the Programme teams are usually setting up a form for people to submit proposals, then ask a few people to vote, and select the higher scores, that would be a simplistic and very incorrect description of what really happens in the Programme teams.
Besides setting up a platform for submitting proposals, there are lots of tasks that are very time-consuming, like:
- Anonymising proposals for reviewers to have a neutral approach when reviewing proposals,
- Setting up reviewers groups and communicating processes and the expected output,
- Interpretation of reviews, to avoid people mistakenly giving an incorrect negative score to proposals,
- Once scored, the submissions needs to be categorized and analyzed to create a balanced schedule which attendees expect,
There are lots of hard decisions to make. We are truthful when communicating that good proposals need to be rejected or left in the waiting list, which brings the next major invisible challenge: cancelled talks.
Every year, we have experienced lots of cancellations of confirmed talks due to many reasons, from VISA problems to no-shows. Yep, you read that right, sometimes we got confirmed speakers not showing up. Many “emergency talks” need to be quickly accepted in order to provide a schedule without many missing talks. This is the most stressful part of running a Programme team, and usually, people are not aware of it.
EP: How has volunteering at EuroPython impacted your own career or learning journey?
The human aspect of the conference is the most important part for me, I truly believe everyone volunteering to conferences wants the best for the event. Through this process, you end up knowing people better, and understand their motivations and decision-making direction. We, humans, are a complicated species, and wherever you have human interactions, you will have agreement and disagreement. Learning how these relationships are developed, and doing your best to avoid too much friction, is a very tricky but fulfilling part of this. I believe this has had a good impact on my career and learning journey, because at my paid work, I have a technical lead/manager position, where similar situations can occur.
Something else I have seen closely, but I have not benefited from so far, is the networking that you get out of volunteering at the conference. If you prove yourself to work well and are looking for a job, I think it’s an amazing opportunity for you to potentially find your next challenge. We have people from many companies around the globe, who certainly will consider you in case they have open positions, more importantly, if you have good dynamics with other volunteers, that works better for you than submitting your resume.
EP: Is there something about the programming community that made you want to give back by participating in EuroPython?
Totally, I remember that one of the motivations I had when starting contributing was wanting to improve things I believed (from my little bubble) needed improvement. Back then at EuroPython 2019 (my first one), I remember feeling very isolated and looking only at a few people that were doing their best to integrate new people into the community. Additionally, I noticed that not many talks were bringing new topics from my perspective, so I thought of maybe helping with reviews.
After speaking at EuroPython 2019, I failed to get a talk accepted in 2020, but decided to join remotely as well, and in 2021 got another talk accepted, but failed again in 2022. Then I thought about participating by volunteering so I could enjoy the conference not only by giving talks, but helping it directly. After all, organizers were doing a call for volunteers after each event, so it was just a matter of time.
Once I started volunteering, I noticed it was a challenging but gratifying experience, so I have been around since then, helping with whatever I can. It’s important to understand that even if you are in charge of a team, you are still a conference volunteer, so running around bringing items, contacting people, or taking care of other volunteers is your responsibility as well.
I enjoy being in touch with friends, having a nice time, and celebrating when the conference is over.
EP: If you could describe the volunteer experience in three words, what would they be?
I’d say for me the words are: “Motivating - Challenging - Rewarding”.
Motivating, because initially you are swamped with ideas on how to improve everything and make an even better conference.
Challenging, because you need to be prepared to solve problems, some of which are new, and it is up to you to do your best.
Rewarding, because of those moments when everything is over, and you see the smile on people’s faces, and how much they are looking forward to the next one. That is a nice reward.
EP: Did you have any unexpected or funny experiences during the EuroPython?
Leaving behind the unexpected last-minute cancellations experiences that I mentioned before, which were very unexpected, I believe that meeting people in-real-life has been very cool! You meet people in online communities all the time, people working on modules or projects that you use, or who you have seen online. Additionally, getting to know people who have been organizing EuroPython in the past or other conferences around Europe and the world has been very nice.
From the funny side, I’d say there are little things, like discovering you attended events together with other folks, but back then you didn’t know each other, or that in the past there was some interaction on Telegram, Discord, or IRC (yeah, I’m that old) and noticing “wow, so you are <nickname>!”.
EP: What keeps you coming back to volunteer year after year?
As I mentioned before, I think the human part is what motivates me the most. I know that by being there, I’m responsible for making things work well, for people to be happy, comfortable, enjoying talks, or talking in the corridor. And maybe some people will think “You know what? Next time I will be volunteering, I love this conference”, this is what brings me to EuroPython every year.
Meeting other volunteers, organizers and attendees that I haven’t met for many months, even years, is always a good excuse as well. EuroPython has become a meeting point for many people in our community, and I would like for that to never disappear.
EP: What is the key to being a good volunteer at EuroPython?
Being responsible for what you committed yourself to do is the key. We all have issues which we need to handle and leave the conference aside, but many people fail to communicate. Believe me, people will understand those situations, but it is very important to be upfront and say “Hey, I don’t have time this week/month, can someone take care of ___?”.
This is volunteering work, but volunteering still needs reliable people, otherwise things can fail. Many people feel bad not being able to do something, but as long as you don’t disappear without a trace, and people don’t need to figure out what you were working on, or if someone else should pick up your tasks, that’s a great portion of what defines “a good volunteer” in my eyes.
EP: Thank you, Cristián!
March 06, 2026
Talk Python to Me
#539: Catching up with the Python Typing Council
You're adding type hints to your Python code, your editor is happy, autocomplete is working great. But then you switch tools and suddenly there are red squiggles everywhere. Who decides what a float annotation actually means? Or whether passing None where an int is expected should be an error? It turns out there's a five-person council dedicated to exactly these questions -- and two brand-new Rust-based type checkers are raising the bar. On this episode, I sit down with three members of the Python Typing Council -- Jelle Zijlstra, Rebecca Chen, and Carl Meyer -- to learn how the type system is governed, where the spec and the type checkers agree and disagree, and get the council's official advice on how much typing is just enough.<br/> <br/> <strong>Episode sponsors</strong><br/> <br/> <a href='https://talkpython.fm/sentry'>Sentry Error Monitoring, Code talkpython26</a><br> <a href='https://talkpython.fm/agentic-ai'>Agentic AI Course</a><br> <a href='https://talkpython.fm/training'>Talk Python Courses</a><br/> <br/> <h2 class="links-heading mb-4">Links from the show</h2> <div><strong>Guests</strong><br/> <strong>Carl Meyer</strong>: <a href="https://github.com/carljm?featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>Jelle Zijlstra</strong>: <a href="https://jellezijlstra.github.io?featured_on=talkpython" target="_blank" >jellezijlstra.github.io</a><br/> <strong>Rebecca Chen</strong>: <a href="https://github.com/rchen152?featured_on=talkpython" target="_blank" >github.com</a><br/> <br/> <strong>Typing Council</strong>: <a href="https://github.com/python/typing-council?tab=readme-ov-file&featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>typing.python.org</strong>: <a href="http://typing.python.org?featured_on=talkpython" target="_blank" >typing.python.org</a><br/> <strong>details here</strong>: <a href="https://github.com/python/typing-council?tab=readme-ov-file#decision-making-considerations" target="_blank" >github.com</a><br/> <strong>ty</strong>: <a href="https://docs.astral.sh/ty/?featured_on=talkpython" target="_blank" >docs.astral.sh</a><br/> <strong>pyrefly</strong>: <a href="https://pyrefly.org?featured_on=talkpython" target="_blank" >pyrefly.org</a><br/> <strong>conformance test suite project</strong>: <a href="https://github.com/python/typing/tree/main/conformance?featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>typeshed</strong>: <a href="https://github.com/python/typeshed?featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>Stub files</strong>: <a href="https://mypy.readthedocs.io/en/stable/stubs.html?featured_on=talkpython" target="_blank" >mypy.readthedocs.io</a><br/> <strong>Pydantic</strong>: <a href="http://pydantic.dev?featured_on=talkpython" target="_blank" >pydantic.dev</a><br/> <strong>Beartype</strong>: <a href="https://github.com/beartype/beartype?featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>TOAD AI</strong>: <a href="https://github.com/batrachianai/toad?featured_on=talkpython" target="_blank" >github.com</a><br/> <strong>PEP 747 – Annotating Type Forms</strong>: <a href="https://peps.python.org/pep-0747/?featured_on=talkpython" target="_blank" >peps.python.org</a><br/> <strong>PEP 724 – Stricter Type Guards</strong>: <a href="https://peps.python.org/pep-0724/?featured_on=talkpython" target="_blank" >peps.python.org</a><br/> <strong>Python Typing Repo (PRs and Issues)</strong>: <a href="https://github.com/python/typing?featured_on=talkpython" target="_blank" >github.com</a><br/> <br/> <strong>Watch this episode on YouTube</strong>: <a href="https://www.youtube.com/watch?v=bzh-0FlAmP0" target="_blank" >youtube.com</a><br/> <strong>Episode #539 deep-dive</strong>: <a href="https://talkpython.fm/episodes/show/539/catching-up-with-the-python-typing-council#takeaways-anchor" target="_blank" >talkpython.fm/539</a><br/> <strong>Episode transcripts</strong>: <a href="https://talkpython.fm/episodes/transcript/539/catching-up-with-the-python-typing-council" target="_blank" >talkpython.fm</a><br/> <br/> <strong>Theme Song: Developer Rap</strong><br/> <strong>🥁 Served in a Flask 🎸</strong>: <a href="https://talkpython.fm/flasksong" target="_blank" >talkpython.fm/flasksong</a><br/> <br/> <strong>---== Don't be a stranger ==---</strong><br/> <strong>YouTube</strong>: <a href="https://talkpython.fm/youtube" target="_blank" ><i class="fa-brands fa-youtube"></i> youtube.com/@talkpython</a><br/> <br/> <strong>Bluesky</strong>: <a href="https://bsky.app/profile/talkpython.fm" target="_blank" >@talkpython.fm</a><br/> <strong>Mastodon</strong>: <a href="https://fosstodon.org/web/@talkpython" target="_blank" ><i class="fa-brands fa-mastodon"></i> @talkpython@fosstodon.org</a><br/> <strong>X.com</strong>: <a href="https://x.com/talkpython" target="_blank" ><i class="fa-brands fa-twitter"></i> @talkpython</a><br/> <br/> <strong>Michael on Bluesky</strong>: <a href="https://bsky.app/profile/mkennedy.codes?featured_on=talkpython" target="_blank" >@mkennedy.codes</a><br/> <strong>Michael on Mastodon</strong>: <a href="https://fosstodon.org/web/@mkennedy" target="_blank" ><i class="fa-brands fa-mastodon"></i> @mkennedy@fosstodon.org</a><br/> <strong>Michael on X.com</strong>: <a href="https://x.com/mkennedy?featured_on=talkpython" target="_blank" ><i class="fa-brands fa-twitter"></i> @mkennedy</a><br/></div>
Peter Bengtsson
logger.error or logger.exception in Python
Consider this Python code:
try:
1 / 0
except Exception as e:
logger.error("An error occurred while dividing by zero.: %s", e)
The output of this is:
An error occurred while dividing by zero.: division by zero
No traceback. Perhaps you don't care because you don't need it.
I see code like this quite often and it's curious that you even use logger.error if it's not a problem. And it's curious that you include the stringified exception into the logger message.
Another common pattern I see is use of exc_info=True like this:
try:
1 / 0
except Exception:
logger.error("An error occurred while dividing by zero.", exc_info=True)
Its output is:
An error occurred while dividing by zero.
Traceback (most recent call last):
File "/Users/peterbengtsson/dummy.py", line 23, in <module>
1 / 0
~~^~~
ZeroDivisionError: division by zero
Ok, now you get the traceback and the error value (division by zero in this case).
But a more convenient function is logger.exception which looks like this:
try:
1 / 0
except Exception:
logger.exception("An error occurred while dividing by zero.")
Its output is:
An error occurred while dividing by zero.
Traceback (most recent call last):
File "/Users/peterbengtsson/dummy.py", line 9, in <module>
1 / 0
~~^~~
ZeroDivisionError: division by zero
So it's sugar for logger.error.
Also, a common logging config is something like this:
import logging
logger = logging.getLogger(__name__)
logging.basicConfig(
format="%(asctime)s - %(levelname)s - %(name)s - %(message)s", level=logging.ERROR
)
So if you use logger.exception what will it print? In short, the same as if you used logger.error. For example, with the logger.exception("An error occurred while dividing by zero.") line above:
2026-03-06 10:45:23,570 - ERROR - __main__ - An error occurred while dividing by zero.
Traceback (most recent call last):
File "/Users/peterbengtsson/dummy.py", line 12, in <module>
1 / 0
~~^~~
ZeroDivisionError: division by zero
Bonus - add_note
You can, if it's applicable, inject some more information about the exception. Consider:
try:
n / 0
except Exception as exception:
exception.add_note(f"The numerator was {n}.")
logger.exception("An error occurred while dividing by zero.")
The net output of this is:
2026-03-06 10:48:34,279 - ERROR - __main__ - An error occurred while dividing by zero.
Traceback (most recent call last):
File "/Users/peterbengtsson/dummy.py", line 13, in <module>
1 / 0
~~^~~
ZeroDivisionError: division by zero
The numerator was 123.
Real Python
Quiz: Python Stacks, Queues, and Priority Queues in Practice
In this quiz, you’ll test your understanding of Python stacks, queues, and priority queues.
You’ll review LIFO and FIFO behavior, enqueue and dequeue operations, and how deques work. You’ll implement a queue with collections.deque and learn how priority queues order elements.
You’ll also see how queues support breadth-first traversal, stacks enable depth-first traversal, and how message queues help decouple services in real-world systems.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Anarcat
Wallabako retirement and Readeck adoption
Today I have made the tough decision of retiring the Wallabako project. I have rolled out a final (and trivial) 1.8.0 release which fixes the uninstall procedure and rolls out a bunch of dependency updates.
Why?
The main reason why I'm retiring Wallabako is that I have completely stopped using it. It's not the first time: for a while, I wasn't reading Wallabag articles on my Kobo anymore. But I had started working on it again about four years ago. Wallabako itself is about to turn 10 years old.
This time, I stopped using Wallabako because there's simply something better out there. I have switched away from Wallabag to Readeck!
And I'm also tired of maintaining "modern" software. Most of the recent commits on Wallabako are from renovate-bot. This feels futile and pointless. I guess it must be done at some point, but it also feels we went wrong somewhere there. Maybe Filippo Valsorda is right and one should turn dependabot off.
I did consider porting Wallabako to Readeck for a while, but there's a perfectly fine Koreader plugin that I've been pretty happy to use. I was worried it would be slow (because the Wallabag plugin is slow), but it turns out that Readeck is fast enough that this doesn't matter.
Moving from Wallabag to Readeck
Readeck is pretty fantastic: it's fast, it's lightweight, everything Just Works. All sorts of concerns I had with Wallabag are just gone: questionable authentication, questionable API, weird bugs, mostly gone. I am still looking for multiple tags filtering but I have a much better feeling about Readeck than Wallabag: it's written in Golang and under active development.
In any case, I don't want to throw shade at the Wallabag folks either. They did solve most of the issues I raised with them and even accepted my pull request. They have helped me collect thousands of articles for a long time! It's just time to move on.
The migration from Wallabag was impressively simple. The importer is well-tuned, fast, and just works. I wrote about the import in this issue, but it took about 20 minutes to import essentially all articles, and another 5 hours to refresh all the contents.
There are minor issues with Readeck which I have filed (after asking!):
- add justified view for articles (Android app)
- more metadata in article display (Android app)
- show the number of articles in the label browser
- ignore duplicates (Readeck will happily add duplicates, whereas Wallabag at least tries to deduplicate articles -- but often fails)
But overall I'm happy and impressed with the result.
I'm also both happy and sad at letting go of my first (and only, so far) Golang project. I loved writing in Go: it's a clean language, fast to learn, and a beauty to write parallel code in (at the cost of a rather obscure runtime).
It would have been much harder to write this in Python, but my experience in Golang helped me think about how to write more parallel code in Python, which is kind of cool.
The GitLab project will remain publicly accessible, but archived, for the foreseeable future. If you're interested in taking over stewardship for this project, contact me.
Thanks Wallabag folks, it was a great ride!
Israel Fruchter
Maybe ORM/ODM are not dead? Yet...
So, let’s pick up where we left off. A couple of weeks ago, I wrote about how I took a 4-year-old fever dream—an ODM for Cassandra and ScyllaDB called coodie—and let an AI build the whole thing while I sipped my morning coffee.
It was a fun experiment. But then, a funny coincidence happened (or maybe the algorithm just has a sick sense of humor).
Right after I hit publish and started feeling good about my newfound “prompt engineer” status, I was listening to an episode of the pythonbytes podcast discussing Michael Kennedy’s recent post, Raw+DC: The ORM pattern of 2026?. The overarching thesis of their discussion? ORMs and ODMs are fundamentally dead. They are a relic of the past, bloated, abstraction-heavy, and ultimately, absolute performance killers.
I actually fired off a Twitter thread in response to it. And honestly, at first, I had to concede. They make a really good point. I spend my days deep in the ScyllaDB trenches, where we fight for every single microsecond. Putting a thick Python abstraction layer on top of a highly optimized driver usually sounds like a brilliant way to turn a sports car into a tractor.
But it got me thinking. How bad was coodie? Was my AI-generated Beanie-wannabe actually a performance disaster waiting to happen?
Giving it a test run
Like any respectable developer looking for an excuse to avoid real work, I decided to put my money where my AI-generated mouth is. Instead of sitting at my desk, I just offloaded the whole task to the Copilot agent from my phone to run some extensive benchmarks.
I didn’t just want to compare coodie to existing solutions like cqlengine. I wanted to establish an absolute performance floor. I wanted to test it against the Raw+DC pattern (Python dataclasses + hand-written CQL with prepared statements) to see exactly how much the “ORM tax” was really costing us.
The test spun up a local ScyllaDB node, hammered it with various workloads—inserts, reads, conditional updates, batch operations—and fetched the results back.
The finally surprising results
I had the agent run the script. I fully expected coodie to be heavily penalized. We all accept slower performance in exchange for autocomplete, declarative schemas, and not writing raw CQL strings.
I stared at the results the agent sent back to my screen. Then I told it to clear the cache, restart the Scylla container, and run it again just to be sure.
The results were genuinely surprising, and running them actually highlighted a few spots where we could squeeze out even more performance, leading straight to PR #190 to apply those lessons learned.
Here is the breakdown of what the agent and I found across the board.
Three-way Benchmark Results (scylla driver)
| Benchmark | Raw+DC (µs) | coodie (µs) | cqlengine (µs) | coodie vs Raw+DC | coodie vs cqlengine |
|---|---|---|---|---|---|
| single-insert | 456 | 485 | 615 | 1.06× | 0.79× ✅ |
| insert-if-not-exists | 1,180 | 1,170 | 1,370 | ~1.00× | 0.85× ✅ |
| insert-with-ttl | 448 | 469 | 640 | 1.05× | 0.73× ✅ |
| get-by-pk | 461 | 520 | 665 | 1.13× | 0.78× ✅ |
| filter-secondary-index | 1,370 | 2,740 | 8,530 | 2.00× 🟠 | 0.32× ✅ |
| filter-limit | 575 | 627 | 1,220 | 1.09× | 0.51× ✅ |
| count | 904 | 1,500 | 1,590 | 1.66× 🟡 | 0.94× ✅ |
| partial-update | 409 | 960 | 542 | 2.35× 🔴 | 1.77× ❌ |
| update-if-condition (LWT) | 1,140 | 1,620 | 1,340 | 1.42× 🟡 | 1.21× ❌ |
| single-delete | 941 | 925 | 1,190 | ~1.00× | 0.78× ✅ |
| bulk-delete | 872 | 921 | 1,200 | 1.06× | 0.77× ✅ |
| batch-insert-10 | 596 | 634 | 1,700 | 1.06× | 0.37× ✅ |
| batch-insert-100 | 42,800 | 1,960 | 52,900 | 0.05× 🚀 | 0.04× ✅ |
| collection-write | 448 | 485 | 679 | 1.08× | 0.71× ✅ |
| collection-read | 478 | 508 | 689 | 1.06× | 0.74× ✅ |
| collection-roundtrip | 939 | 1,060 | 1,380 | 1.13× | 0.77× ✅ |
| model-instantiation | 0.671 | 2.02 | 12.1 | 3.01× 🔴 | 0.17× ✅ |
| model-serialization | 10.1 | 2.05 | 4.56 | 0.20× 🚀 | 0.45× ✅ |
Summary of Findings
Near Parity on the Hot Paths: For standard reads (get-by-pk), inserts, deletes, and basic limited queries, coodie is operating with a completely negligible overhead compared to writing raw CQL by hand (usually hovering around 5-13% tax). It also outperforms cqlengine across the board on these operations.
Pydantic V2 is a beast: We need to remember that Pydantic V2 is basically a blazing-fast Rust engine wearing a Python trench coat. The data validation, object instantiation, and serialization happen at the speed of Rust, keeping the overhead minimal from the start. Just look at the serialization and batch performance multipliers!
The AI kept it lean: The code the LLM generated wasn’t building massive ASTs or doing unnecessary query translations. coodie essentially formats a clean dictionary and hands it straight to the underlying Scylla driver to do its native magic. It gets out of the way.
Lessons Learned (PR #190): Even with great initial numbers, running the full benchmark suite revealed bottlenecks on things like partial-update and count. We realized we were doing redundant data validation during read operations when fetching data we already knew was valid straight from the database. By bypassing the extra validation pass and loading the raw rows more directly into the Pydantic models (using model_construct() for DB data), we can shave off the remaining overhead.
Wrapping up
So, maybe ORM/ODM are not dead? Yet…
If the final overhead of using an ODM on hot paths is a measly 5-13%, but in exchange I get full type-safety, declarative schemas, and zero boilerplate, I am taking the ODM every single time.
It seems my digital sidekick built something that is actually production-ready, and with a little bit of human-driven optimization, it screams.
You can check out the code, star it, or run your own tests over at github.com/fruch/coodie.
PRs are welcome. יאללה, let’s see how fast we can make it.
March 05, 2026
The Python Coding Stack
You Store Data and You Do Stuff With Data • The OOP Mindset
Learning how to use a tool can be challenging. But learning why you should use that tool–and when–is sometimes even more challenging.
In this post, I’ll discuss the OOP mindset–or, let’s say, one way of viewing the object-oriented paradigm. I debated with myself whether to write this article here on The Python Coding Stack. Most articles here are aimed at the “intermediate-ish” Python programmer, whatever “intermediate” means. Most readers may feel they already understand the ethos and philosophy of OOP. I know I keep discovering new perspectives from time to time. So here’s a perspective I’ve been exploring in courses I ran recently.
As you can see, I decided to write this post. At worst, it serves as revision for some readers, perhaps a different route towards understanding why we (sometimes) define classes and (always) use objects in Python. And beginners read these articles, too!
If you feel you’re an OOP Pro, go ahead and skip this post. Or just read it anyway. It’s up to you. You can always catch up with anything you may have missed from The Python Coding Stack’s Archive instead – around 120 full-length articles and counting!
This post is inspired by the introduction to OOP in Chapter 7 in The Python Coding Book – The Relaxed and Friendly Programming Book for Beginners
Meet Mark, a market seller. He sets up a stall in his local market and sells tea, coffee, and home-made biscuits (aka cookies for those in North America).
I’ll write two and a half versions of the code Mark could use to keep track of his stock and sales. The first one–that’s the half–is not very pretty. Don’t write code like this. But it will serve as a starting point for the main theme of this post. The second version is a stepping stone towards the third.
First (Half) Version • Not Pretty
Mark is also learning Python in his free time. He starts writing some code:
He was planning to write some functions to update the stock, change the cost price and selling price, and deal with a sale from his market stall.
But he stopped.
We’ll also stop here with this version.
Mark is a Python beginner, but even he knew this was not the best way to store the relevant data for each product he sells.
Four separate objects? Not ideal. Mark knows that the data in these four lists are linked to each other. The first items in each list belong together, and so on. But he’ll need to manually ensure he maintains these links in the code he writes. Not impossible, but it’s asking for trouble. So many things can go wrong. And he’ll have a tough time writing the code, making all those links in every line he writes.
Mark knows what the link is between the various values. But the computer program doesn’t. The computer program sees four separate data structures, unrelated to each other. You can try writing an update_stock() function with this version to see the challenges, the manual work needed to connect data across separate structures.
Let’s move on.
Second Version • Dictionary and Functions
Mark learnt about dictionaries. They’re a great way to group data together:
Now there’s one data structure that contains the three products he sells. This structure contains three other data structures, each containing all the relevant data for each product.
The data are structured into these dictionaries to show what belongs where. In this version, the Python program “knows” which data items belong together. Each dictionary contains related values.
Well done, Mark! He understood the need to organise the data into a sensible structure. This is not the only combination of nested data structures Mark could use, but this is a valid option. Certainly better than the first version!
He can write some functions now. First up is update_stock():
Let’s ignore the fact that Mark is accessing the global variable products within the function. We’ll have a chat with Mark about this.
Still, there’s now a function called update_stock(). It needs the product name as an argument. It also needs the stock amount to increase (or decrease if the value is negative).
Let’s look at another function Mark wrote:
This function also needs the product name as one of its arguments. It also needs a second argument: the quantity sold.
Mark wrote other functions, but I’ll keep this section brief so I won’t show them. However, many of Mark’s functions have a few things in common:
They require the product name as one of the arguments. This makes sense since the operations Mark needs to carry out depend on which product he’s dealing with.
They make changes to the data in one of the inner dictionaries defined within
products.Some of them return data. Others don’t.
Great. Mark is happy with his effort.
He structured the data sensibly so it’s well organised. Separately, he wrote functions that use data from those data structures–the inner dictionaries in products and sometimes make changes to the data in those data structures.
Your call…
The Python Coding Place offers something for everyone:
• a super-personalised one-to-one 6-month mentoring option
$ 4,750
• individual one-to-one sessions
$ 125
• a self-led route with access to 60+ hrs of exceptional video courses and a support forum
$ 400
Which The Python Coding Place student are you?
Third Version • Class and Objects
Mark’s code stores data. The nested dictionaries in products deal with the storage part. His code also does stuff* with the data through the functions he wrote.
*stuff is not quite a Python technical term, in case you’re wondering. But it’s quite suitable here, I think!
Lots of programs store data and do stuff with data.
Object-oriented programming takes these two separate tasks–storing data and doing stuff with data–and combines them into a single “unit”. In OOP, this “unit” is the object. Objects are at the centre of the OOP paradigm, which is why OOP is called OOP, after all!
Mark’s progression from the first version to the second relied on structuring the data into units–the nested dictionary structure.
The progression from the second version to the third takes this a step further and structures the data and the functions that act on those data into a single unit: the object.
This structure means that the data for each object is contained within the object, and the actions performed are also contained within that object. Everything is self-contained within the object.
Let’s build a Product class for Mark. But let’s do this step by step, using the code from the second version and gradually morphing it into object-oriented code. This will help us follow the transition.
First, let’s create the class:
Nothing much to see so far. But this is the shell that will contain all the instructions for creating this “unit,” including all the relevant data and functionality.
You can already create an instance of this class – that’s another way of saying an object created from this class:
Whereas Product represents the class, Product() is an instance of the class. Note the parentheses. There’s only one Product, but you can create many instances using Product() .
Bundling In the Functionality • Methods
Now, let me take a route I recently explored for the first time while teaching OOP in a beginners’ course. It’s only subtly different from my “usual” teaching route, but I think it helped me appreciate the topic from a distinct perspective. So, here we go.
Let’s copy and paste the functions from version two directly into this class. Warning: This won’t work. We’ll need to make some changes. But let’s use this as a starting point:
To keep you on your toes, we change the terminology here. These are functions. However, when they’re defined within a class definition, we call them methods. But they’re still functions. Everything you know about functions applies equally to methods.
You’ve seen that each function in version two needed to know which product it was dealing with. That’s why the first parameter is product_name. You used product_name to fetch the correct values when you needed the selling price, the cost price, or the number of items in stock.
However, now that we’re in the OOP domain, the object will contain all the data it needs–you’ll add the data-related code soon. You don’t need to fetch the data from anywhere else, just from the object itself.
Therefore, the functions defined within the class – the methods – no longer need the product name as the first argument. Instead, they need the entire object itself since this object contains all the data the method needs about the object.
We just need a parameter name to describe the object itself. How about self?
The method signatures now include self as the first parameter and whatever else the methods need as the remaining parameters.
Incidentally, although you can technically name this first parameter anything you want, there’s a strong convention to always use self. So it’s best to stick with self!
The methods are the tools you use to bundle functionality into the object. Let’s pause on working on the methods for now and shift our attention to how to store the data, which also needs to be bundled into the object.
Bundling In the Data • Data Attributes
Let’s get back to the code that creates an instance of the Product class:
The expression Product() does a few things behind the scenes. First, it creates a new blank object. Then it initialises this object. To initialise the object, Python needs to “do stuff”. Therefore, it needs a method. But not just any method. A special method:
You can’t choose the name of this method. It must be .__init__(). But it’s still a method. It still takes self as the first parameter since it still needs access to the object itself. This method creates variables that are attached to the object. You can think of the dot as attaching name to self and so on for the others. We tend not to call these variables – more new terminology just for the OOP paradigm – they’re data attributes. They’re object attributes that store data.
For now, these data attributes contain default values: the empty string for the .name data attribute and 0 for the others. However, when you create an object, you often want to supply some or all of the data that the object needs. Often, you want to pass this information when you create the object:
However, when you run the code, you get an error:
Traceback (most recent call last):
...
a_product = Product(”Coffee”, 1.98, 3.2, 100)
TypeError: Product.__init__() takes 1 positional
argument but 5 were givenThe error message mentions Product.__init__(). Note how you don’t explicitly use .__init__() in the expression to create a Product object. However, Python calls this special method behind the scenes. And when it does, it complains about the arguments you passed:
"Product.__init__() takes 1 positional argument..."is the first part of the error message. Makes sense, since you haveselfas the one and only parameter within the definition of the.__init__()special method."...but 5 [arguments] were given", the error message goes on to say. Wait, why 5? You pass four objects when you createProduct(): the string"Coffee", the floats1.98and3.2, and the integer100. Python can’t count, it seems?
Not quite. When Python calls a method, it automatically passes the object as the first argument. You don’t see this. You don’t need to do anything about it. It happens behind the scenes. So, soon after Product() creates a blank new instance, it calls .__init__() and passes the object as the first argument to .__init__(), the one that’s assigned to the parameter self.
Recall that methods are functions. So, Python automatically passes the object as the first argument so that these functions (methods) have access to the object.
That’s why the error message says that five arguments were passed: the object itself and the four remaining arguments.
This tells you that .__init__() needs five parameters. The first is self, which is the first parameter in these methods. Let’s add the remaining four:
The code no longer raises an error since the number of arguments passed to Product() when you create the object – including the object itself, which is implied – matches the number of parameters in .__init__(). However, you want to shift the data into the data attributes you created earlier. These data attributes are the storage devices attached to the object. You want the data to be stored there:
The parameters are used only to transfer the data from the method call to the data attributes. From now on, the data are stored in the data attributes, which are attached to the object.
Note that the data attribute names don’t have to match the parameter names. But why bother coming up with different names? Might as well use the same ones!
Now, you can create any object you wish, each having its own data:
Here’s the output:
Coffee
Tea
3.2
2.25Every instance of the Product class will have a .name, .cost_price, .selling_price, and .stock. But each instance will have its own values for those data attributes. Each object is distinct from the others. The data is self-contained within the object.
Back to the Functionality • Methods
Let’s look at the class so far, which still has code pasted from version two earlier:
Let’s focus on .update_stock() first. Note how in my writing style guide, I use a leading dot when writing method names, such as .update_stock(). That’s because they’re also attributes of the object. To call a method, you call it through the object, such as a_product.update_stock(30).
Recall that Python will pass the object itself as the first argument to the method. That’s the argument assigned to self. You then pass 30 to the parameter stock_increment.
But this means that this method can only act on an object that already exists, that already has the data attributes it needs. You no longer need to check whether the product exists. If you’re calling this method, you’re calling it on a product. Checks on existence will happen elsewhere in your code.
So, all that’s left is to increment the stock for this object. The current stock is stored in the .stock data attribute – the data attribute that’s connected to the object. But you passed the object to .update_stock(), so you can access self.stock from within the method:
And that’s it. Everything is self-contained within the object. The method acts on the object and modifies one of the object’s data attributes.
How about the .sell() method?
Again, you can remove the validation to check that the product exists. A method is called on a Product object, so it exists. This method then uses three of the object’s data attributes to perform its task.
Note that you can add similar validation in .update_stock() to ensure the stock doesn’t dip below 0. However, stock_increment can be negative to enable reducing the stock. You could even use .update_stock() within .sell() if you wish.
Ah, what about the biscuits? You can still use a list of products, but this time the list contains Product objects:
Or, if you prefer, you can use a dictionary:
Let’s trial out this dictionary and the Product class:
In this basic use case, you use the item input by the user to fetch the corresponding Product object from the products dictionary. You assign this object to the variable name product. Then you can call its methods, such as .sell(), and access its data attributes, such as .name and .stock.
Here’s the output from this code, including sample user inputs:
Enter item: coffee
Enter quantity of Coffee sold: 3
The income from this sale is £9.60
The profit from this sale is £3.66
Remaining Coffee units: 97Final Words
The aim of this article is not to provide a comprehensive and exhaustive walkthrough of OOP. I wrote elsewhere about OOP. You can start from The Python Coding Book and then read the seven-part series A Magical Tour Through Object-Oriented Programming in Python • Hogwarts School of Codecraft and Algorithmancy.
You can learn about the syntax and the mechanics of defining and using classes. And that’s important if you want to write classes. But just as importantly, you need to adopt an OOP mindset. A central point is how OOP bundles data and functionality into a single unit, the object, and how everything stems from that structure.
In this post, you saw how data is bundled into the object through data attributes. And you bundled the high-level functionality that matters for your object through the methods .update_stock() and .sell(). Mark will need more of these methods, as you can imagine.
However, the object also includes low-level functionality. What should Python do when it needs to print the object? How about if it needs to add it to another object? Should that be possible? Should the object be considered False under any circumstance, say? These operations are defined by an object’s special methods, also known as dunder methods. I proposed thinking of these as “plumbing methods” recently: “Python’s Plumbing” Is Not As Flashy as “Magic Methods” • But Is It Better?
Therefore, there’s plenty of high-level and low-level functionality bundled within the object. And the data, of course.
Here’s another relatively recent post you may enjoy in case you missed it when it was published: My Life • The Autobiography of a Python Object.
In summary, OOP structures data and functionality into a single unit – the object. Then, your code is oriented around this object.
Do you want to master Python one article at a time? Then don’t miss out on the articles in The Club which are exclusive to premium subscribers here on The Python Coding Stack
Photo by gomed fashion
Code in this article uses Python 3.14
The code images used in this article are created using Snappify. [Affiliate link]
Join The Club, the exclusive area for paid subscribers for more Python posts, videos, a members’ forum, and more.
You can also support this publication by making a one-off contribution of any amount you wish.
For more Python resources, you can also visit Real Python—you may even stumble on one of my own articles or courses there!
Also, are you interested in technical writing? You’d like to make your own writing more narrative, more engaging, more memorable? Have a look at Breaking the Rules.
And you can find out more about me at stephengruppetta.com
Further reading related to this article’s topic:
Appendix: Code Blocks
Code Block #1
products = ["Coffee", "Tea", "Biscuits"]
cost_price = [1.98, 0.85, 3.1]
selling_price = [3.2, 2.25, 5]
stock = [100, 200, 25]
Code Block #2
products = {
"Coffee": {
"cost_price": 1.98,
"selling_price": 3.2,
"stock": 100,
},
"Tea": {
"cost_price": 0.85,
"selling_price": 2.25,
"stock": 200,
},
"Biscuits": {
"cost_price": 3.1,
"selling_price": 5,
"stock": 25,
},
}
Code Block #3
# ...
def update_stock(product_name, stock_increment):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
products[product_name]["stock"] += stock_increment
# We'll need to have a chat with Mark about the issues
# with accessing global variables from functions.
# Let's let it pass here!
Code Block #4
# ...
def sell(product_name, quantity):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
if quantity <= 0:
raise ValueError("Quantity must be greater than zero")
if products[product_name]["stock"] < quantity:
raise ValueError("Not enough stock")
products[product_name]["stock"] -= quantity
selling_price = products[product_name]["selling_price"]
cost_price = products[product_name]["cost_price"]
income = selling_price * quantity
profit = (selling_price - cost_price) * quantity
return income, profit
Code Block #5
class Product:
...
Code Block #6
class Product:
...
a_product = Product()
Code Block #7
class Product:
# Just pasting the functions from Version 2 here for now.
# This still doesn't work. It's a work-in-progress
def update_stock(product_name, stock_increment):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
products[product_name]["stock"] += stock_increment
# We'll need to have a chat with Mark about the issues
# with accessing global variables from functions.
# Let's let it pass here!
def sell(product_name, quantity):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
if quantity <= 0:
raise ValueError("Quantity must be greater than zero")
if products[product_name]["stock"] < quantity:
raise ValueError("Not enough stock")
products[product_name]["stock"] -= quantity
selling_price = products[product_name]["selling_price"]
cost_price = products[product_name]["cost_price"]
income = selling_price * quantity
profit = (selling_price - cost_price) * quantity
return income, profit
Code Block #8
class Product:
def update_stock(self, stock_increment):
# rest of code pasted from version two (for now)
def sell(self, quantity):
# rest of code pasted from version two (for now)
Code Block #9
# ...
a_product = Product()
Code Block #10
class Product:
# We'll make improvements to this method soon
def __init__(self):
self.name = ""
self.cost_price = 0
self.selling_price = 0
self.stock = 0
Code Block #11
# ...
a_product = Product("Coffee", 1.98, 3.2, 100)
Code Block #12
class Product:
def __init__(self, name, cost_price, selling_price, stock):
self.name = ""
self.cost_price = 0
self.selling_price = 0
self.stock = 0
Code Block #13
class Product:
def __init__(self, name, cost_price, selling_price, stock):
self.name = name
self.cost_price = cost_price
self.selling_price = selling_price
self.stock = stock
Code Block #14
# ...
a_product = Product("Coffee", 1.98, 3.2, 100)
another_product = Product("Tea", 0.85, 2.25, 200)
print(a_product.name)
print(another_product.name)
print(a_product.selling_price)
print(another_product.selling_price)
Code Block #15
class Product:
def __init__(self, name, cost_price, selling_price, stock):
self.name = name
self.cost_price = cost_price
self.selling_price = selling_price
self.stock = stock
# Just pasting the functions from Version 2 here for now.
# This still doesn't work. It's a work-in-progress
def update_stock(self, stock_increment):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
products[product_name]["stock"] += stock_increment
# We'll need to have a chat with Mark about the issues
# with accessing global variables from functions.
# Let's let it pass here!
def sell(self, quantity):
if product_name not in products:
raise KeyError(f"Product {product_name} not found")
if quantity <= 0:
raise ValueError("Quantity must be greater than zero")
if products[product_name]["stock"] < quantity:
raise ValueError("Not enough stock")
products[product_name]["stock"] -= quantity
selling_price = products[product_name]["selling_price"]
cost_price = products[product_name]["cost_price"]
income = selling_price * quantity
profit = (selling_price - cost_price) * quantity
return income, profit
Code Block #16
# ...
def update_stock(self, stock_increment):
self.stock += stock_increment
Code Block #17
# ...
def sell(self, quantity):
if quantity <= 0:
raise ValueError("Quantity must be greater than zero")
if self.stock < quantity:
raise ValueError("Not enough stock")
self.stock -= quantity
income = self.selling_price * quantity
profit = (self.selling_price - self.cost_price) * quantity
return income, profit
Code Block #18
# ...
products = [
Product("Coffee", 1.98, 3.2, 100),
Product("Tea", 0.85, 2.25, 200),
Product("Biscuits", 3.1, 5, 25),
]
Code Block #19
products = {
"Coffee": Product("Coffee", 1.98, 3.2, 100),
"Tea": Product("Tea", 0.85, 2.25, 200),
"Biscuits": Product("Biscuits", 3.1, 5, 25),
}
Code Block #20
# ...
# I've kept this demo code brief and simple, so it lacks
# verification and robustness. You can add this if you wish!
item = input("Enter item: ").title()
qty = int(input(f"Enter quantity of {item} sold: "))
product = products[item]
income, profit = product.sell(qty)
print(f"The income from this sale is £{income:.2f}")
print(f"The profit from this sale is £{profit:.2f}")
print(f"Remaining {product.name} units: {product.stock}")
For more Python resources, you can also visit Real Python—you may even stumble on one of my own articles or courses there!
Also, are you interested in technical writing? You’d like to make your own writing more narrative, more engaging, more memorable? Have a look at Breaking the Rules.
And you can find out more about me at stephengruppetta.com
Real Python
Quiz: Spyder: Your IDE for Data Science Development in Python
In this quiz, you’ll test your understanding of Spyder: Your IDE for Data Science Development in Python.
By working through this quiz, you’ll revisit how to install Spyder, explore variables with the Variable Explorer, visualize data with the Plots pane, and profile code to find performance bottlenecks.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Quiz: How to Use the OpenRouter API to Access Multiple AI Models via Python
In this quiz, you’ll test your understanding of How to Use the OpenRouter API to Access Multiple AI Models via Python.
By completing this quiz, you’ll review how OpenRouter provides a unified routing layer, how to call multiple providers from a single Python script, how to switch models without changing your code, and how to compare outputs.
It also reinforces practical skills for making API requests in Python, handling authentication, and processing responses. For deeper guidance, review the tutorial linked above.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Daniel Roy Greenfeld
Top Terminal Tools
When I sit down to code, these are the tools I use at this time whenever I touch code. In alphabetical order:
Atuin
Atuin is a replacement for the default shell history. It saves your history to a local, encrypted SQLite database. Then it allows for blazing fast searches. You can sync your history across devices, and is has a lot of other features. I can't imagine using a terminal without Atuin.
For OSX users, I recommend installing following these instructions.
bat
A Rust-based cat replacement. It has syntax highlighting, line numbers, and a lot of other features that make it a great tool for quickly looking at files in the terminal.
Ghostty
Ghostty is a fast, feature-rich, and cross-platform terminal emulator that I believe works everywhere. Yes, TMUX (and competitors) are more configurable and have more features, but Ghostty just works out of the box. Ghostty eschews the arcane key combinations of its predecessors in favor of intuitive keybindings. A ghostty terminal can be split horizontally and vertically and copy/paste works as expected. It also doesn't appear to interfere with any other shell tool, something that annoyed me about TMUX.
Usually I have three vertical panels:
- On the far left a panel for running general shell commands like git, uv, and whatever. Atuin is a must for me, giving me shell history on steriods so I don't have to remember commands anymore
- In the middle I run a CLI-based text editor, these days usually pad
- On the righthand side a panel for running LLM CLI tools, usually AMP, Codex, and sometimes Claude
GitHub CLI
Even though I use a fraction of the available commands, this is just a really useful tool for working on GitHub-hosted projects.
Pad
Pad is an easy-to-use terminal editor I created for those of us who aren't into vim. It's powered by Will McGugan's Textual. I did what I can to make pad work with typical VS Code keybindings.
Right now I'm not accepting general contributions to Pad. I am still recovering from a head injury and that unfortunately makes reviewing PRs challenging. As I get better I plan to put more features into the project and perhaps even accept contributions. For now it does what I need it to do and is a handy addition to my tool chain.
Various LLM CLI tools
I have no loyalty to any particular LLM provider. I use whatever tool gets the job done and for which I have free or affordable credits. For now I use:
- Amp
- ChatGPT Codex
- Claude Code
For a while I was using gemini-cli, but it stopped working for me weeks ago. Rather than try to debug it I just used the competition.
scikit-learn
Update on array API adoption in scikit-learn
Note: this blog post is a cross-post of a Quansight Labs blog post.
The Consortium for Python Data API Standards developed the Python array API standard to define a consistent interface for array libraries, specifing core operations, data types, and behaviours. This enables ‘array-consuming’ libraries (such as scikit-learn) to write array-agnostic code that can be run on any array API compliant backend. Adopting array API support in scikit-learn means that users can pass arrays from any array API compliant library to functions that have been converted to be array-agnostic. This is useful because it allows users to take advantage of array library features, such as hardware acceleration, most notably via GPUs.
Indeed, GPU support in scikit-learn has been of interest for a long time - 11 years ago, we added an entry to our FAQ page explaining that we had no plans to add GPU support in the near future due to the software dependencies and platform specific issues it would introduce. By relying on the array API standard, however, these concerns can now be avoided.
In this blog post, I will provide an update to the array API adoption work in scikit-learn, since it’s initial introduction in version 1.3 two years ago. Thomas Fan’s blog post provides details on the status when array API support was initially added.
Current status
Since the introduction of array API support in version 1.3 of scikit-learn, several key developments have followed.
Vendoring array-api-compat and array-api-extra
Scikit-learn now vendors both
array-api-compat and
array-api-extra.
array-api-compat is a wrapper around common array libraries (e.g., PyTorch,
CuPy, JAX) that bridges gaps to ensure compatibility with the standard. It
enables adoption of backwards incompatible changes while still allowing array
libraries time to adopt the standard slowly. array-api-extra provides array
functions not included in the standard but deemed useful for array-consuming
libraries.
We chose to vendor these now much more mature libraries in order to avoid the complexity of conditionally handling optional dependencies throughout the codebase. This approach also follows precedent, as SciPy also vendors these packages.
Array libraries supported
Scikit-learn currently supports CuPy ndarrays, PyTorch tensors (testing against all devices: ‘cpu’, ‘cuda’, ‘mps’ and ‘xpu’) and NumPy arrays. JAX support is also on the horizon. The main focus of this work is addressing in-place mutations in the codebase. Follow PR #29647 for updates.
Beyond these libraries, scikit-learn also tests against array-api-strict, a
reference implementation that strictly adheres to the array API specification.
The purpose of array-api-strict is to help automate compliance checks for
consuming libraries and to enable development and testing of array
API functionality without the need for GPU or other specialized hardware.
Array libraries that conform to the standard and pass the array-api-tests suite
should be accepted by scikit-learn and SciPy, without any additional modifications
from maintainers.
Estimators and metrics with array API support
The full list of metrics and estimators that now support array API can be
found in our
Array API support
documentation page. The majority of high impact metrics have now been
converted to be array API compatible. Many transformers are also now
supported, notably LabelBinarizer which is widely used internally and
simplifies other conversions.
Conversion of estimators is much more complicated as it often involves
benchmarking different variations of code or consensus gathering on
implementation choices. It generally requires many months of work by several
maintainers. Nonetheless, support for LogisticRegression, GaussianNB,
GaussianMixture, Ridge (and family: RidgeCV, RidgeClassifier,
RidgeClassifierCV), Nystroem and PCA has been added. Work on
GaussianProcessRegressor is also underway (follow at
PR #33096).
Handling mixed array namespaces and devices
scikit-learn takes a unique approach among ‘array-consuming’ libraries by supporting mixed array namespace and device inputs. This design choice enables the framework to handle the practical complexities of end-to-end machine learning pipelines.
String-valued class labels are common in classification tasks and enable users to work with interpretable categories rather than integer codes. NumPy is currently the only array library with string array support, meaning that any workflow involving both GPU-accelerated computation and string labels necessarily involves mixed array type inputs.
Mixed array input support also enables flexible pipeline workflows. Pipelines
provide significant value by chaining preprocessing steps and estimators into
reusable workflows that prevent data leakage and ensure consistent
preprocessing. However, they have an intentional design limitation: pipeline
steps can transform feature arrays (X) but cannot modify target arrays
(y). Allowing mixed array inputs means a pipeline can include a
FunctionTransformer step that moves feature data from CPU to GPU to leverage
hardware acceleration, while allowing the target array, which cannot be
modified, to remain on CPU.
For example, mixed array inputs enable a pipeline where string classification
features are encoded on CPU (as only NumPy supports string arrays), converted
to torch CUDA tensors, then passed to the array API-compatible
RidgeClassifier for GPU-accelerated computation:
from functools import partial
from sklearn.linear_model import RidgeClassifier
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import FunctionTransformer, TargetEncoder
pipeline = make_pipeline(
# Encode string categories with average target values
TargetEncoder(),
# Convert feature array `X` to Torch CUDA device
FunctionTransformer(partial(torch.asarray, dtype="float32", device="cuda"))
RidgeClassifier(solver="svd"),
)
Work on adding mixed array type inputs for metrics and estimators is underway and expected to progress quickly. This work includes developing a robust testing framework, including for pipelines using mixed array types (follow PR #32755 for details).
Finally, we have also revived our work to support the ability to fit and predict on different namespaces/devices. This allows users to train models on GPU hardware but deploy predictions on CPU hardware, optimizing costs and accommodating different resource availability between training and production environments. Follow PR #33076 for details.
Challenges
The challenges of array API adoption remain largely unchanged from when this work began. These are also common to other array-consuming libraries, with a notable addition: the need to handle array movement between namespaces and devices to support mixed array type inputs.
Array API Standard is a subset of NumPy’s API
The array API standard only includes widely-used functions implemented across most array libraries, meaning many NumPy functions are absent. When such a function is encountered while adding array API support, we have the following options:
- add the function to
array-api-extra- this allows other array-consuming libraries to benefit and allows sharing of maintenance burden, but is only relevant for more widely used functions - add our own implementation in scikit-learn - these functions live in
sklearn/utils/_array_api.py - check if SciPy implements an array API compatible version of the function
The quantile function illustrates this decision-making process. quantile
is not included in the standard as it is not widely used (outside of
scikit-learn) and while it is implemented in most array libraries, the
set of quantile methods supported and their APIs vary. Currently, scikit-learn maintains its
own array API compatible version that supports both weights and NaNs, but due
to the maintenance burden we decided to investigate alternatives. SciPy has an
array API compatible implementation, but it did not support weights. We thus
investigated adding quantile to array-api-extra; however, during this
effort, SciPy decided to add weight support. Thus, we ultimately decided to
transition to the SciPy implementation once our minimum SciPy version allows.
Compiled code
Many performance-critical parts of scikit-learn are written using compiled code extensions in Cython, C or C++. These directly access the underlying memory buffers of NumPy arrays and are thus restricted to CPU.
Metrics and estimators, with compiled code, handle this in one of two ways:
convert arrays to NumPy first or maintain two parallel branches of code, one
for NumPy (compiled) and one for other array types (array API compatible).
When performance is less critical or array API conversion provides no gains
(e.g., confusion_matrix), we convert to NumPy. When performance gains are
significant, we accept the maintenance burden of dual code paths. This was the case for
LogisticRegression and the extensive process required for making such implementation
decisions can be seen in the
PR #32644.
Unspecified behaviour in the standard
The array API standard intentionally leaves some function behaviors
unspecified, permitting implementation differences across array libraries. For
example, the order of unique elements is not specified for the unique_*
functions and as of NumPy version 2.3, some unique_* functions no longer
return sorted values. This will require code amendments in cases where sorted
output was relied upon.
Similarly, NaN handing is also unspecified for sort; however, in this case, all
array libraries currently supported by scikit-learn follow NumPy’s NaN
semantics, placing NaNs at the end. This consistency eliminates the need for
special handling code, though comprehensive testing remains essential when
adding support for new array libraries.
Device transfer
Mixed array namespace and device inputs necessitates conversion of arrays between different namespaces and devices. This presented a number of considerations and challenges.
The array API standard adopted DLPack as the recommended data interchange protocol. This protocol is widely implemented in array libraries and offers an efficient, C ABI compatible protocol for array conversion. While this provided us with an easy way to implement these transfers, there were limitations. Cross-device transfer capability was only introduced in DLPack v1, released in September 2024. This meant that only the latest PyTorch and CuPy versions have support for DLPack v1. Moreover, not all array libraries have adopted support yet. We therefore implemented a ‘manual’ fallback; however, this requires conversion via NumPy when the transfer involves two non-NumPy arrays. Additionally, there are no DLPack tests in array-api-tests, a testing suite to verify standard compliance, leaving DLPack implementation bugs easier to overlook. Despite these challenges, scikit-learn will benefit from future improvements, such as addition of a C-level API for DLPack exchange that bypasses Python function calls, offering significant benefit for GPU applications.
Beyond the technical considerations, there were also user interface considerations. How should we inform users that these conversions, which incur memory and performance cost, are occurring? We decided against warnings, which risk being ignored or becoming a nuisance, and to instead clearly document this behaviour. Additionally, different devices have different data type limitations; for example, Apple MPS only supports float32. How best to handle these differences when performing conversions while ensuring users are informed of precision impacts is an ongoing consideration.
A quick benchmark
Array API support for Ridge regression was added in version 1.5, enabling
GPU-accelerated linear models in scikit-learn. Combined with support of
several transformers, this allows for complete preprocessing and estimation
pipelines on GPU.
The following benchmark shows the use of the MaxAbsScaler transformed
followed by Ridge regression using randomly generated data with 500,000
samples and 300 features. The benchmarks were run on AMD Ryzen Threadripper
2970WX CPU, NVIDIA Quadro RTX 8000 GPU and Apple M4 GPU (Metal 3).
The figure below shows the performance speed up on CuPy, Torch CPU and Torch GPU relative to NumPy.
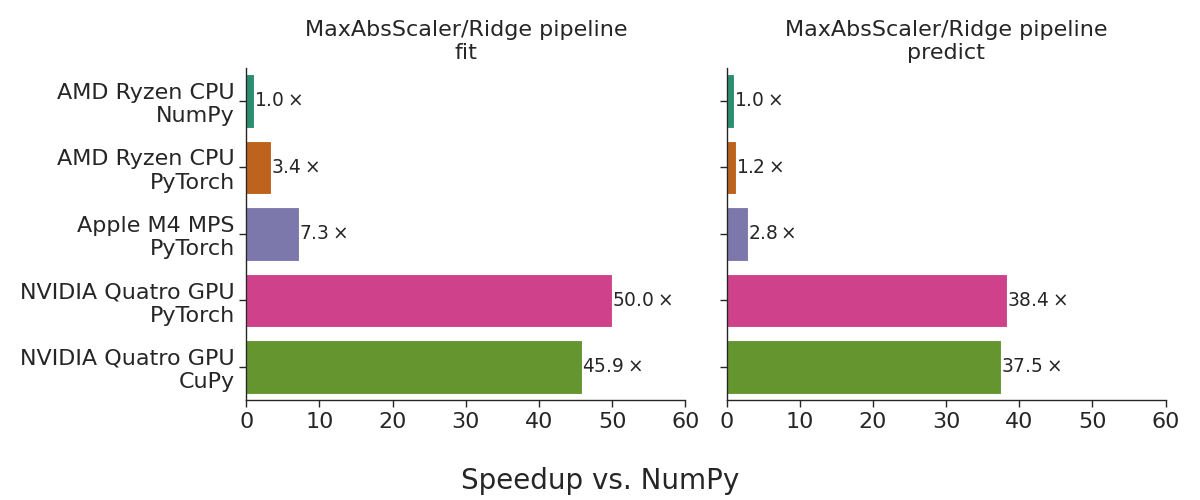
Performance speedup relative to NumPy across different backends.
The observed speedups are representative of performance gains achievable with sufficiently large datasets on datacenter-grade GPUs for linear algebra-intensive workloads. Mobile GPUs, such as those in laptops, would typically yield more modest improvements.
Note that scikit-learn’s Ridge regressor currently only supports ‘svd’
solver. We selected this solver for initial implementation as it exclusively
uses standard-compliant functions available across all backends and is the
most stable solver. Support for the ‘cholesky’ solver is also underway (see
details in PR #29318).
Looking forward
As of version 1.8, array API support is still in experimental mode and thus not enabled by default. However, we welcome early adopters and interested users to try it and report any issues. See our documentation for details on enabling array API support.
Before removing experimental status, we would like to:
- develop a system for automatically documenting functions and classes that support array API, potentially with the ability to add relevant details
- mixed array type input support
- support fit and predict on different hardware by allowing conversion of fitted estimators between namespaces/devices using utility functions
- improved testing, in particular for the new mixed array type functionalities
- improved documentation, including adding an example to our gallery
- decide on the minimal dependency versions required
- get real world user feedback
Alongside these infrastructure and framework improvements, we look forward to adding support for more estimators. These improvements will deliver production-ready GPU support and flexible deployment options to scikit-learn users. We welcome community involvement through testing and feedback throughout this development phase.
Acknowledgements
Work on array API in scikit-learn has been a combined effort from many contributors. This work was partly funded by CZI and NASA Roses.
I would like to thank Olivier Grisel, Tim Head and Evgeni Burovski for helping me with my array API questions.
Armin Ronacher
AI And The Ship of Theseus
Because code gets cheaper and cheaper to write, this includes re-implementations. I mentioned recently that I had an AI port one of my libraries to another language and it ended up choosing a different design for that implementation. In many ways, the functionality was the same, but the path it took to get there was different. The way that port worked was by going via the test suite.
Something related, but different, happened with chardet. The current maintainer reimplemented it from scratch by only pointing it to the API and the test suite. The motivation: enabling relicensing from LGPL to MIT. I personally have a horse in the race here because I too wanted chardet to be under a non-GPL license for many years. So consider me a very biased person in that regard.
Unsurprisingly, that new implementation caused a stir. In particular, Mark Pilgrim, the original author of the library, objects to the new implementation and considers it a derived work. The new maintainer, who has maintained it for the last 12 years, considers it a new work and instructs his coding agent to do precisely that. According to author, validating with JPlag, the new implementation is distinct. If you actually consider how it works, that’s not too surprising. It’s significantly faster than the original implementation, supports multiple cores and uses a fundamentally different design.
What I think is more interesting about this question is the consequences of where we are. Copyleft code like the GPL heavily depends on copyrights and friction to enforce it. But because it’s fundamentally in the open, with or without tests, you can trivially rewrite it these days. I myself have been intending to do this for a little while now with some other GPL libraries. In particular I started a re-implementation of readline a while ago for similar reasons, because of its GPL license. There is an obvious moral question here, but that isn’t necessarily what I’m interested in. For all the GPL software that might re-emerge as MIT software, so might be proprietary abandonware.
For me personally, what is more interesting is that we might not even be able to copyright these creations at all. A court still might rule that all AI-generated code is in the public domain, because there was not enough human input in it. That’s quite possible, though probably not very likely.
But this all causes some interesting new developments we are not necessarily ready for. Vercel, for instance, happily re-implemented bash with Clankers but got visibly upset when someone re-implemented Next.js in the same way.
There are huge consequences to this. When the cost of generating code goes down that much, and we can re-implement it from test suites alone, what does that mean for the future of software? Will we see a lot of software re-emerging under more permissive licenses? Will we see a lot of proprietary software re-emerging as open source? Will we see a lot of software re-emerging as proprietary?
It’s a new world and we have very little idea of how to navigate it. In the interim we will have some fights about copyrights but I have the feeling very few of those will go to court, because everyone involved will actually be somewhat scared of setting a precedent.
In the GPL case, though, I think it warms up some old fights about copyleft vs permissive licenses that we have not seen in a long time. It probably does not feel great to have one’s work rewritten with a Clanker and one’s authorship eradicated. Unlike the Ship of Theseus, though, this seems more clear-cut: if you throw away all code and start from scratch, even if the end result behaves the same, it’s a new ship. It only continues to carry the name. Which may be another argument for why authors should hold on to trademarks rather than rely on licenses and contract law.
I personally think all of this is exciting. I’m a strong supporter of putting things in the open with as little license enforcement as possible. I think society is better off when we share, and I consider the GPL to run against that spirit by restricting what can be done with it. This development plays into my worldview. I understand, though, that not everyone shares that view, and I expect more fights over the emergence of slopforks as a result. After all, it combines two very heated topics, licensing and AI, in the worst possible way.
March 04, 2026
PyCharm
Cursor is now available as an AI agent inside JetBrains IDEs through the Agent Client Protocol. Select it from the agent picker, and it has full access to your project.
If you’ve spent any time in the AI coding space, you already know Cursor. It has been one of the most requested additions to the ACP Registry.
What you get
Cursor is known for its AI-native, agentic workflows. JetBrains IDEs are valued for deep code intelligence – refactoring, debugging, code quality checks, and the tooling professionals rely on at scale. ACP brings the two together.
You can now use Cursor’s agentic capabilities directly inside your JetBrains IDE – within the workflows and features you already use.
A growing open ecosystem
Cursor joins a growing list of agents available through ACP in JetBrains IDEs. Every new addition to the ACP Registry means you have more choice – while still working inside the IDE you already rely on. You get access to frontier models from major providers, including OpenAI, Anthropic, Google, and now also Cursor.
This is part of our open ecosystem strategy. Plug in the agents you want and work in the IDE you love – without getting locked into a single solution.
Cursor is focused on building the best way to build software with AI. By integrating Cursor with JetBrains IDEs, we’re excited to provide teams with powerful agentic capabilities in the environments where they’re already working.
– Jordan Topoleski, COO at Cursor
Get started
You need version 2025.3.2 or later of your JetBrains IDE with the AI Assistant plugin enabled. From there, open the agent selector, select Install from ACP Registry…, install Cursor, and start working. You don’t need a JetBrains AI subscription to use Cursor as an AI agent.
The ACP Registry keeps growing, and many agents have already joined it – with more on the way. Try it today with Cursor and experience agent-driven development inside your JetBrains IDE. For more information about the Agent Client Protocol, see our original announcement and the blog post on the ACP Agent Registry support.
Python Morsels
Invent your own comprehensions in Python
Python doesn't have tuple, frozenset, or Counter comprehensions, but you can invent your own by passing a generator expression to any iterable-accepting callable.
Table of contents
Generator expressions pair nicely with iterable-accepting callables
Generator expressions work really nicely with Python's any and all functions:
>>> numbers = [2, 1, 3, 4, 7, 11, 18]
>>> any(n > 1 for n in numbers)
True
>>> all(n > 1 for n in numbers)
False
In fact, I rarely see any and all used without a generator expression passed to them.
Note that generator expressions are made with parentheses:
>>> (n**2 for n in numbers)
<generator object <genexpr> at 0x74c535589b60>
But when a generator expression is the sole argument passed into a function:
>>> all((n > 1 for n in numbers))
False
The double set of parentheses (one to form the generator expression and one for the function call) can be turned into just a single set of parentheses:
>>> all(n > 1 for n in numbers)
False
This special allowance was added to Python's syntax because it's very common to see generator expressions passed in as the sole argument to specific functions.
Note that passing generator expressions into iterable-accepting functions and classes makes something that looks a bit like a custom comprehension. Every iterable-accepting function/class is a comprehension-like tool waiting to happen.
Tuple comprehensions
Python does not have tuple …























 Author:
Author:
 Lucy Liu
Lucy Liu